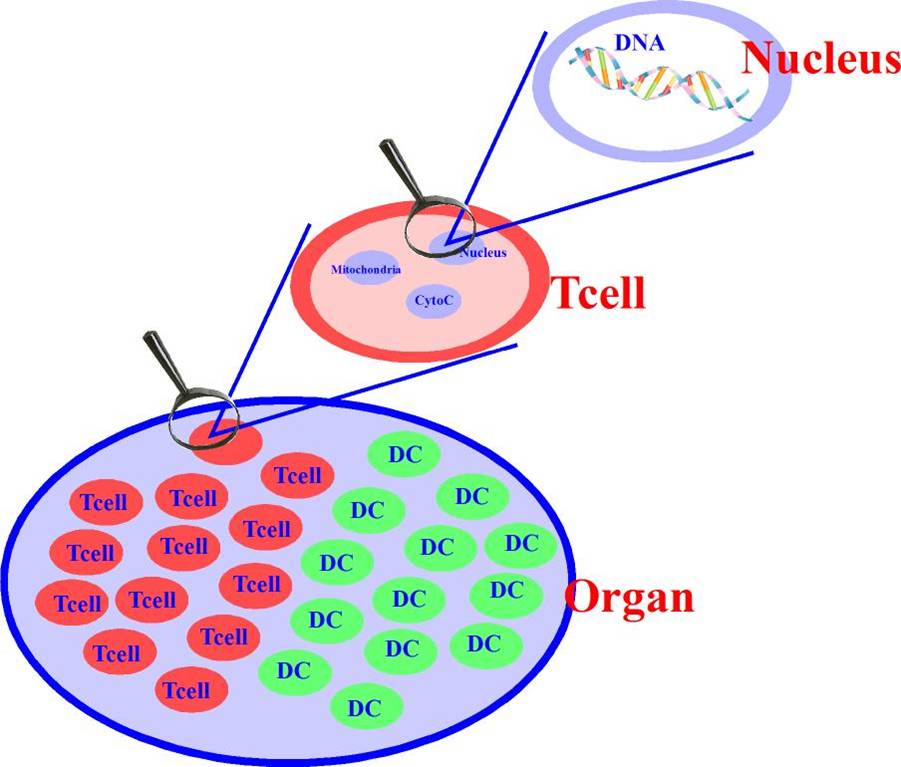
Structural complexity includes molecules, cells and organs. Cells evolve, differentiate, proliferate, and communicate with other cells. Organs are of higher complexity and consist of a variety of cell populations. Molecules are the messengers in the system and facilitate the evolution of cell states. This structural hierarchy has a very intuitive and simple representation in our framework, which is implemented as Agent Based Models involving hierarchical agents. The objects may also have a location if needed for a particular modeling requirement, as in the case of migration of cells from a tissue to a lymph node, or to the blood. Objects communicate through surface-surface interactions (via surface molecules), through the secretion of molecules from one object into another, and more. Object, such as cells, may migrate between higher level compartments, representing for example organs, the blood, the lymphatic system, etc. This is facilitated using chemokines and their receptors as in the real system.
In our mathematical model the hierarchical structure is represented as follows:
First, there are simple objects, such as molecules, genes, processes (i.e. CDx, TLRx, TNF, Caspase-x).
Simple objects are described in terms of their identity, i.e., a name, and abundance which attains one of the discrete values allowed (none, low, high, very high, etc.). Their evolution in the system is governed by a set of reactions as explained later. Simple objects may also correspond to biological processes which are either not fully understood, or that the user has decided to represent in a coarse way. Examples are high level processes in a cell such as Cell Division, Apoptosis, or DNA damage. They can be used in this modeling environment in a transparent way, affecting and being affected by molecular or other cellular processes.
Next, there are complex objects (referred as objects), are made up of simple objects, and other complex objects, such as cells (i.e. Macrophage, Neutrophil, B cell, T cell, D cell, Nucleus).
Objects are described in terms of their surface molecules and internal objects which themselves may be complex, i.e., mitochondria, nucleus, etc.
Syntax for defining an object is given in the following form:
<name> =: ( <SimpleObject List>,( <Object List>)),
where <SimpleObject List> is a set of names of simple objects together with their abundance, and <Object List> is a set of other objects residing inside this one. Objects communicate through surface-surface interactions (via surface molecules), through the secretion of molecules from one object into another, and more. Objects, such as cells, may migrate between higher level compartments that represent organs, the blood, the lymphatic system, etc. This is facilitated using chemokines and their receptors as in reality.
Finally, a population of these complex objects form organs (i.e. Lung, Liver, etc.).
Object Tree. In CS terminology, objects can be viewed as trees where the nodes of the tree are containers for ‘simple objects’, and the children of a given node represent its internal objects, see models C-E. The rule regarding interactions is: ‘Simple objects’ can interact with other ‘simple objects’ in the same node, in the parent node, and in all sibling nodes.